Got milk? Let Your Genetic Profile Dictate That
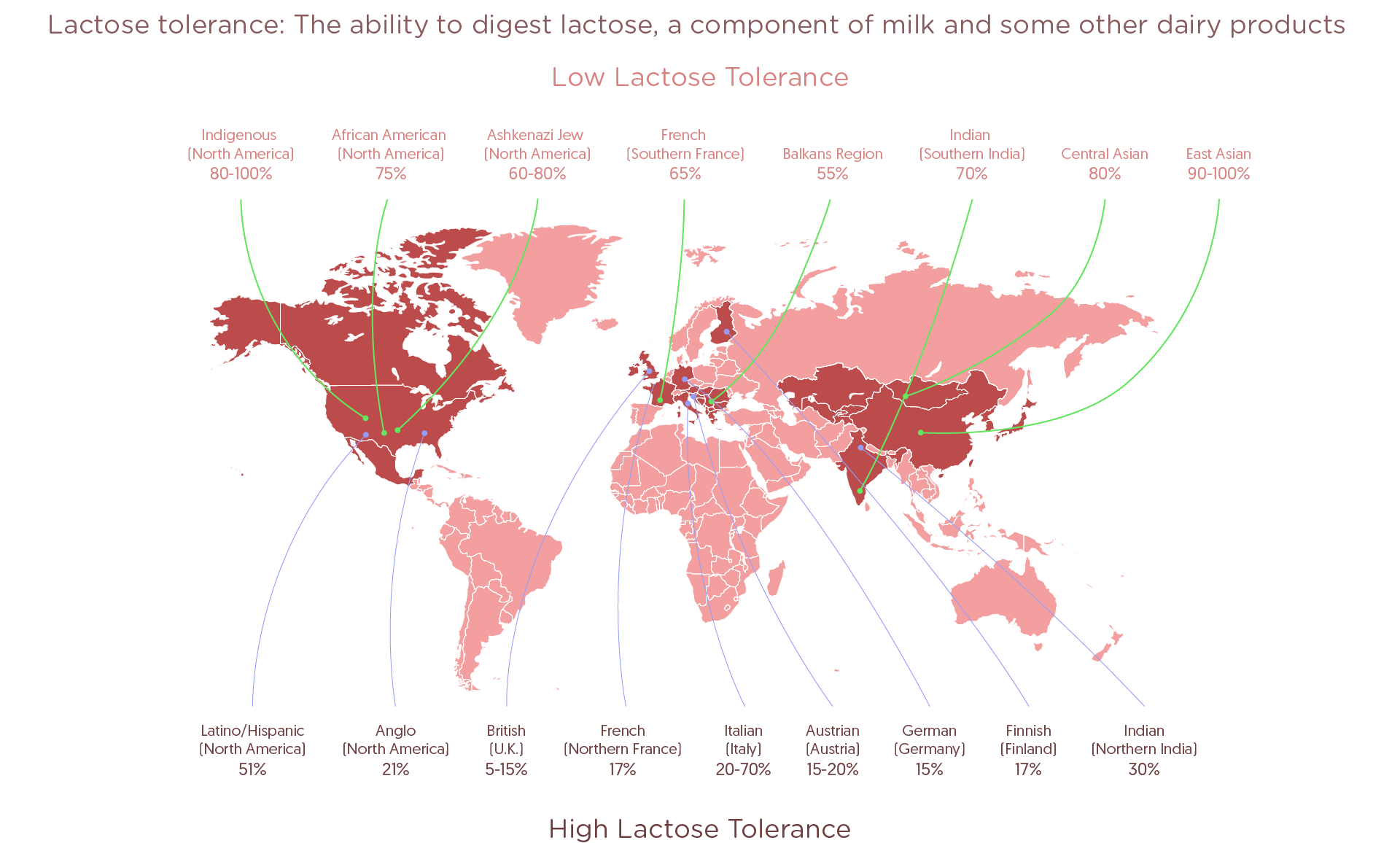
In the United States and several other countries milk is aplenty, and yet not everyone can enjoy it. Following the consumption of a tall glass of milk, 75% of African Americans and American Indians, 10% of Africa’s Tutsi tribe, 50% of Spanish and French people and 99% of Chinese will have to endure consequent stomach upset and other rather unpleasant digestive difficulties. The culprit is lactose, milk’s primary sugar.
Most adults in the world cannot digest lactose and as such are said to be lactose intolerant.
Babies, however, grow up on milk without any problems for the most part. So given that most of us grew up happily drinking milk from a bottle or breast – what happened in the intervening time? Why is it that so many babies enjoy lactose and adults avoid it?
Research reveals that European ancestors from the Stone Age had the same issues – they couldn’t digest milk either. How did they make the transition from milk-a-phobics to milkaholics? The answer is in genetic evolution that takes us from today all the way back to thousands of years ago, when it all began.
Evolution from Milk-a-Phobics to Milkaholics
Milk has got protein, a lot of micronutrients, calcium and carbohydrates – it is pretty nutritious. In a way, it was like a kind of superfood for the ancient Neolithic farmer. And after it is processed into cheese or yoghurt – or after some of the lactose was removed – even those who were lactose intolerant could indirectly benefit from milk.
Today, an adult’s ability to digest milk seems like an obvious benefit, but that has not always been the case. Turns out that lactose tolerance is only beneficial in environments and cultures where individuals have access to domesticated dairy animals.
Most babies have the ability to digest milk without having to endure stomach issues thanks to an enzyme called lactase. However, up until several years ago that enzyme shut down once an individual reached adulthood, implicating that most adults were lactose intolerant. Scientists refer to this as ‘lactase nonpersistent.’
For the majority of Northern and Central Europeans as well as certain African and Middle Eastern populations, that is not the case any longer. It is estimated that the development of lactose tolerance took nearly 20,000 years along with strong selective pressure.
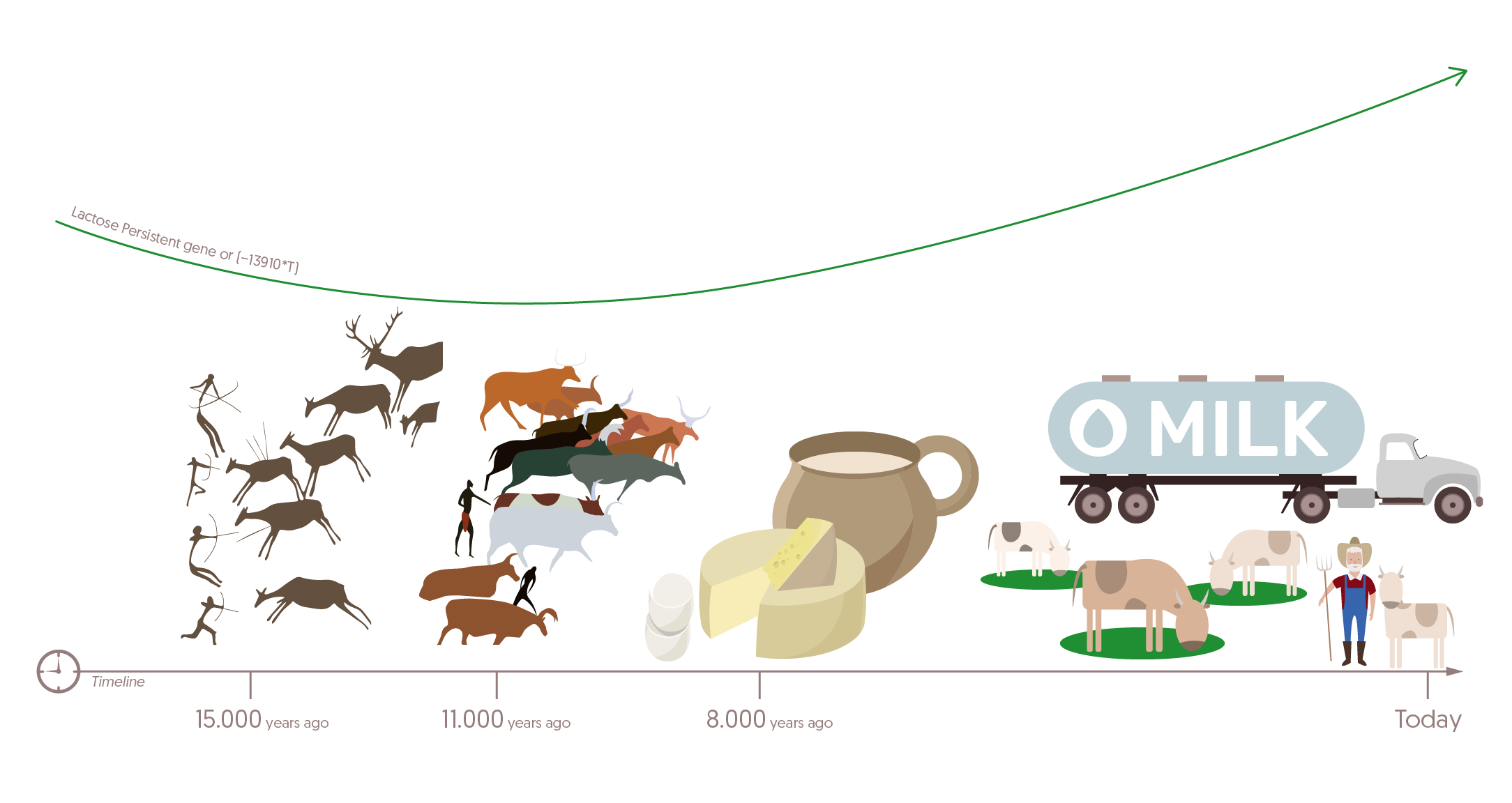
Approximately 11,000 years ago, farming started to replace hunting and gathering in the Middle east. This drove cattle herders to make cheese or yoghurt via milk fermentation – in other words, to reduce lactose amounts in dairy products to tolerable levels [1].
Several thousand years ago, presumably as a result of the aforementioned, Europeans were able to produce lactase and therefore drink milk throughout their lives all due to a mutation that spread across the continent.
As such, difficult conditions that resulted in harvest failures were not lethal – people had milk they could ingest and digest and therefore survive nutritional hardships.
In what is now Turkey, humans started to milk newly domesticated cows, goats and sheep around 8000 years ago. It is exactly at that time that mutations near the gene responsible for lactase production became more frequent. Not surprisingly, adult lactose tolerance developed at that time, as well. And although estimates vary, the mutation responsible for that dates back to between 2,000 and 20,000 years [2].
Scientists have yet to solidify the reason behind why lactose tolerance evolved so quickly.
The current running theory is that as more of our ancestors drank milk, those who were lactose tolerant were able to survive and therefore pass on the lactose tolerance genes to their offspring. That still begs the chicken-and-egg question, however, of which came first: was it dairy herding that was adopted by those who were able to digest milk, or did the advent of dairy herding favor those individuals who had the genetic predisposition to digest it, thereby spreading it to subsequent generations?
Joachim Burger at the University of Mainz speculates that milk-drinking mutations were uncommon before the practice of dairy. Following humans learning the practice of herding cattle, the mutation spread quickly as those who had it were at an advantage [3].
Furthermore, Tishkoff and Reed also suspect that in addition to the extra calories that were gained from milk digestion, lactose persistence may also have allowed individuals with the mutation to stay alive during unfavorable conditions such as droughts [4,5].
However, the LeCHE ((Lactase Persistence in the early Cultural History of Europe) are still trying to decipher the exact reasons behind the advantages of milk drinking. One assumption is that when people moved north, milk provided a hedge against famine as milk products could be stored for longer in colder climates, providing nutrition independently of growing seasons or bad harvests [1]. There are models that were designed by LeCHE members that speculate that, as Middle Eastern Neolithic cultures moved into Europe, farming and herding technologies helped them out-compete the local hunter-gatherers. “And as the southerners pushed north, the LP allele ‘surfed’ the wave of migration,” according to Gerbault, a population geneticist at University Collee London.
As Neolithic farmers settled in parts of southern Europe before the mutation appeared, lactase persistence had a harder time becoming established in those regions [1,6]. But the agricultural society expanded towards the northern and western regions into new territories, allowing lactase persistence to have a big impact. “As the population grows quickly at the edge of the wave, the allele can increase in frequency,” says Gerbault.
The aforementioned patterns are still visible today – lactase persistence is rare in southern Europe (40% of Greek and Turkish populations), while in Britain and in Scandinavia, more than 90% of adults have the ability to digest milk.
Genes/SNPS
The small intestine is lined with cells, called intestinal epithelial cells that have finger-like projections called microvilli. These cells take nutrients from food as it passes through the intestine so that these can get into the bloodstream. Groups of these microvilli are also known as the brush border, where lactase breaks down lactose into glucose and galactose from milk and other dairy products. The two are sugars that are ready to be absorbed.
The LCT gene is responsible for directing the making of lactase. LCT mutations are believed to interfere with the functioning of lactase.
In other words, infants, who are otherwise born without any LCT mutations and that are lactose intolerant are said to have a LCT mutation that shuts down lactase. It is believed that this mutation gradually decreases in activity or expression in most humans and therefore causes intolerance in adulthood.
So if lactase is controlled by LCT, is there anything that controls the gene itself? A DNA sequence called a regulatory element (also referred to as a DNA replication licensing factor) that is located near another gene, MCM6, controls the expression of LCT. In other words, those of us who have inherited changes in this element that result in continuous lactase production in the small intestine have the superpower to digest lactose throughout life (they’ve got milk). However, those (unfortunate ones) among us without those changes have less of an ability to digest lactose as they get older. This results in symptoms of lactose intolerance (think abdominal pain, bloating, cramping, diarrhea, nausea, vomiting) [7].
In more detail, the MCM6 gene codes for the highly conserved mini-chromosome maintenance proteins (MCM), a group known as helicase. What takes place next is that these helicases adhere to specific areas of DNA and temporarily unwind the DNA helices and so it enables each strand or helix to be copied. In other words, MCM6 is essential for the start of genome replication or ‘making more DNA’ [8,9,10].
So how are MCM6 and LCT related exactly? MCM6 contains two regions that regulate LCT expression, and mutations in these regulatory regions, or areas that control LCT function, manifest themselves as what we call lactose tolerance in adult life [11, 12].
So this may sound repetitive, but to reiterate, here is how lactose intolerance comes to be. There are at least 4 SNPs in the MCM6 gene that have an effect on LCT expression, all of which allow a person to degrade lactose into adulthood. Although the word ‘mutation’ sounds bad, it is not. These tiny mutations are actually good for you because they let you enjoy your glass of milk without the uncomfortable symptoms mentioned above. Those who do not have the mutations cannot break down milk into sugars and are considered lactose intolerant.
One last note about genetic variants and lactose persistence (lactose tolerance).
While the majority of other polymorphisms (genetic variations) are referred to as ‘loss-of-function’ mutations, literally meaning that the mutation causes a loss in a physical function, the LP mutations are gain-of-function mutations. This means that one’s ability to break down lactose into sugars is gained. Therefore, while loss-of-function mutations lead to dietary restrictions (they tell us what we can and cannot eat), gain-of-function mutations pretty much tell you to keep on with certain food choices.
Applying This Information to YOU
Whether you have experienced negative side effects or discomforts following dairy intake, it would be beneficial to know whether you are lactose intolerant or not. This, again, falls into the category of investigating one’s genetic background in order to personalize and optimize a dietary regimen which eventually leads to a better quality of life.
Certainly, the fields of nutrigenetics and nutrigenomics must be given honorary mention – without them, we would not be aware of the genetic underpinnings that drive our dietary choices. But what do these ‘omics’ terms mean? Broadly speaking, they are opposites. Nutrigenetics seeks to highlight how gene variants affect the responses to diet. Nutrigenomics, on the other hand, wants to know how our food choices affect genetic expression. Suffice it to say that both fall under the umbrella of nutritional genomics whose aim is to advance health through the application of personalized nutrition [1].
Want to know if you’ve got milk?
If you have already conducted genetic testing or you are interested in finding out more about nutrigenetics and nutrigenomics and how they apply to you, if you find this field interesting and/or if you simply would like to remain informed, please visit us at www.ginihealth.com and subscribe to our blog for regular updates.
References:
- Curry, A. (2013, August). Archaeology: The milk revolution. Nature, 500(7460), pp. 20-22.
- Leonardi, M., Gerbault, P., Thomas, M. G., & Burger, J. (2012). The evolution of lactase persistence in Europe. A synthesis of archaeological and genetic evidence. International Dairy Journal, 22(2), 88-97.
- Ancient DNA solves milk mystery. (2007, February 26). Retrieved from: https://www.nature.com/news/2007/070226/full/news070226-4.html.
- Check, E. (2006, December). How Africa learned to love the cow. Nature, 444(21), pp. 994-996.
- Tishkoff, S. A., Reed, F. A., Ranciaro, A., Voight, B. F., Babbitt, C. C., Silverman, J. S., Powell, K., Mortensen, H. M., Hirbo, J. B., Osman, M., Ibrahim, M., Omar, S. A., Lema, G., Nyambo, T. B., Ghori, J., Bumpstead, S., Pritchard, J. K., Wray, G. A., and Deloukas, P. (2007). Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genetics 39:31-40. (5)
- Burger, J., Kirchner, M., Bramanti, B., Haak, W., and Thomas, M. G. (2007). Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proceedings of the National Academy of Sciences USA 104(10):3736-3741.
- Smith, G. D., Lawlor, D. A., Timpson, N. J., Baban, J., Kiessling, M., Day, I. N., & Ebrahim, S. (2009). Lactase persistence-related genetic variant: population substructure and health outcomes. European journal of human genetics, 17(3), 357.
- Enattah, N. S., Jensen, T. G., Nielsen, M., Lewinski, R., Kuokkanen, M., Rasinpera, H., ... & Natah, A. (2008). Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. The American Journal of Human Genetics, 82(1), 57-72.
- Ingram, C. J., Mulcare, C. A., Itan, Y., Thomas, M. G., & Swallow, D. M. (2009). Lactose digestion and the evolutionary genetics of lactase persistence. Human genetics, 124(6), 579-591.
- Itan, Y., Jones, B. L., Ingram, C. J., Swallow, D. M., & Thomas, M. G. (2010). A worldwide correlation of lactase persistence phenotype and genotypes. BMC evolutionary biology, 10(1), 36.
- Enattah, N. S., Sahi, T., Savilahti, E., Terwilliger, J. D., Peltonen, L., & Järvelä, I. (2002). Identification of a variant associated with adult-type hypolactasia. Nature genetics, 30(2), 233.
- Mattar, R., de Campos Mazo, D. F., & Carrilho, F. J. (2012). Lactose intolerance: diagnosis, genetic, and clinical factors. Clinical and experimental gastroenterology, 5, 113.